::::::::: research ::::::::
Overview
Plants and seeds are the main dietary source of essential nutrient metals, such as zinc (Zn), iron (Fe), manganese (Mn), and copper (Cu). However, plant-based products are also the main entry point for toxic elements, like cadmium (Cd), arsenic (As), mercury (Hg), and lead (Pb). In humans, detrimental effects of toxic metals have been linked to diabetes, hypertension, myocardial infarction, diminished lung function, and certain types of cancer.
Understanding the molecular mechanisms by which plants mobilize and accumulate heavy metals, essential or not, will have two major impacts on human health. First, it will enhance the nutritional value and safety of plant products by ensuring the accumulation of essential metals while avoiding the retention of toxic metals. Second, the identification of genes and molecular mechanisms that allow plants to tolerate and accumulate toxic metals will facilitate the engineering of plants for bioremediation purposes.
Our lab has three main areas of interest:
- Long-distance transport of heavy metals.
- Plant transcriptional responses to environmental stresses.
- Metabolic engineering and crop improvement.
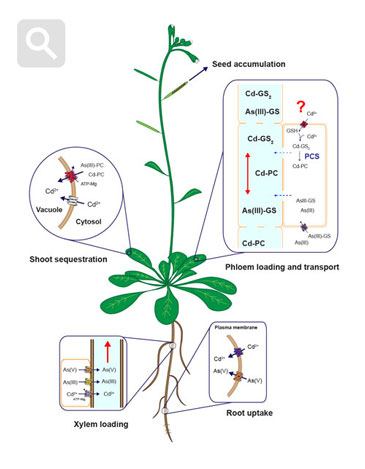
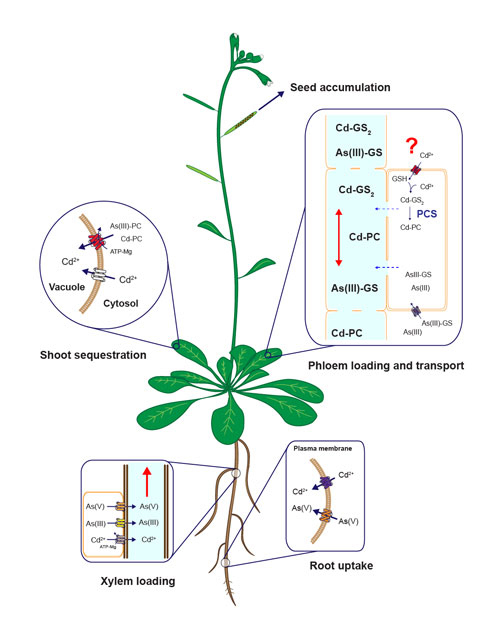
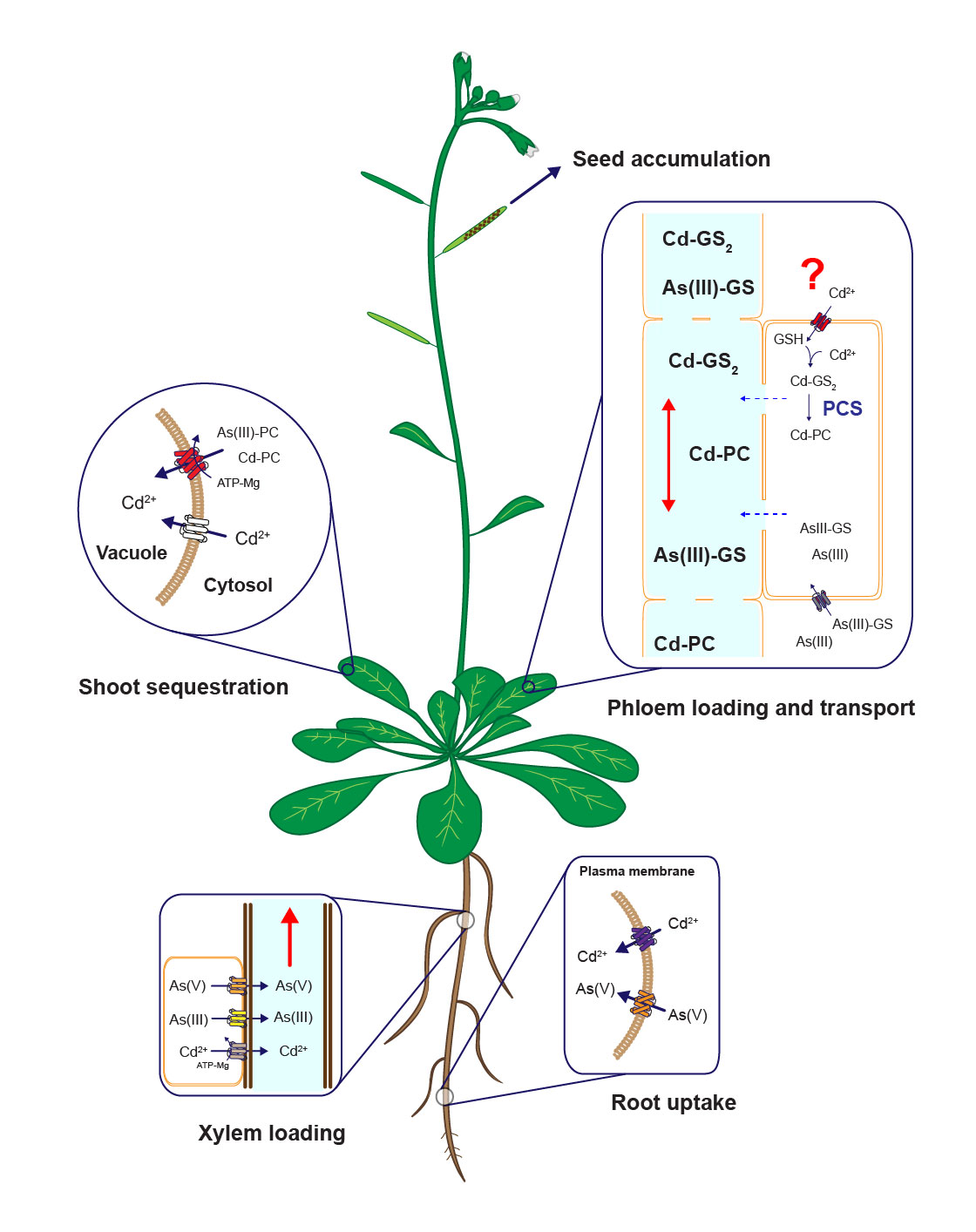
Diagram of uptake, sequestration and long-distanc cadmium and arsenic tarnsport mechanisms in plants. Modified from Mendoza-Cózatl et al., 2011.
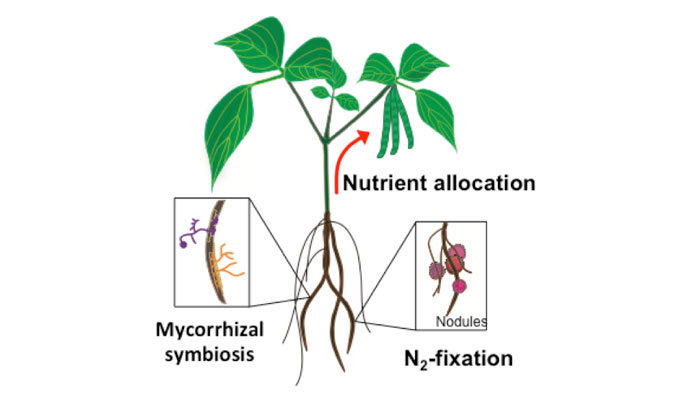
Long-distance transport of heavy metals
Distribution of heavy metals between roots and shoots is a dynamic process orchestrated by several plasma membrane transporters, metal-chelating molecules, xylem-loading/unloading, and phloem-loading/unloading processes. Root-to-shoot transport of metals occurs mainly through the xylem; however, due to the limited transpiration rate within reproductive tissues, the xylem plays only a minor role in allocating nutrients into the seeds. Phloem transport, on the other hand, plays a key role in delivering nutrients, including heavy metals, to developing seeds. Our lab is pursuing the isolation, identification, and characterization of phloem-specific transporters of trace metals (essential and toxic) using soil and hydroponic systems, radiotracer imaging and elemental profiling (ionomics).

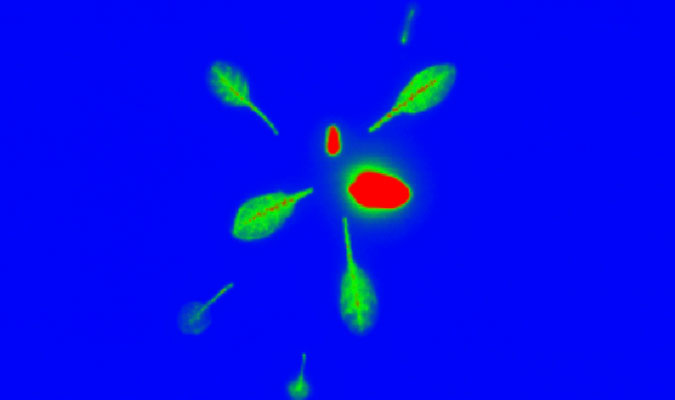
above Arabidopsis plants grown using a hydroponic system.
below radio tracer imaging of Arabidopsis.
Plant transcriptional responses to environmental stresses
Cells have evolved sophisticated mechanisms to sense, respond and adapt to changes in their surrounding environment. Perception of individual stimuli relies on specific receptors and signaling cascades that are activated within seconds, leading to changes in protein function, metabolism and gene expression. These signaling cascades and their downstream effects (e.g. changes in gene expression) require interactions between proteins within specific networks; therefore, identifying protein networks and sub-networks has been one of the latest and perhaps greatest challenges in biology. Our lab is combining transcriptomics and proteomics at cell-specific resolution to identify the molecular mechanisms by which plants sense and respond to changes in the concentration of essential metals such as iron (Fe) and (Zn). Our lab is also pursuing experiments to identify the mechanisms by which plants detoxify toxic non-essential elements such as Cd and As.
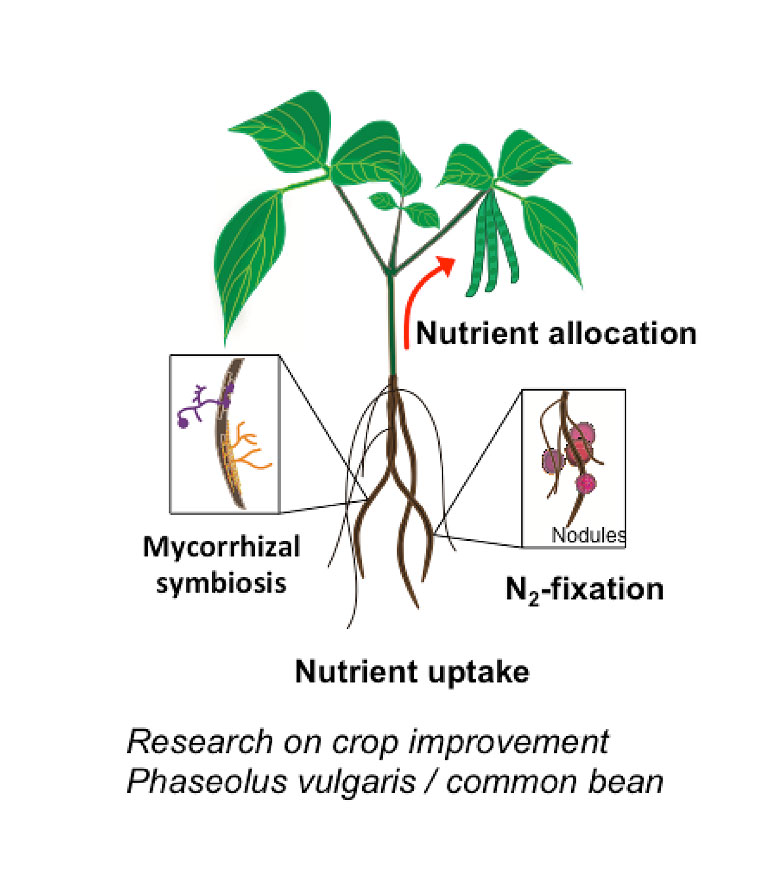
Metabolic engineering and crop improvement
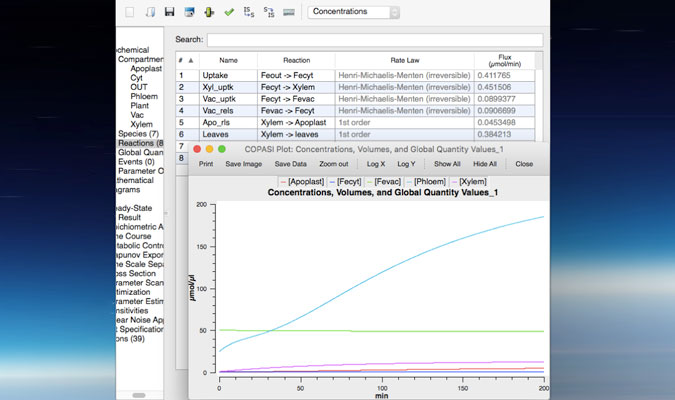
Our lab uses the reference plant Arabidopsis thaliana to unravel the molecular mechanism regulating trace metal homeostasis in plants. Our goal is to learn from Arabidopsis to later transfer this knowledge to crops such as soybean and common bean to improve the nutritional composition of seeds (Fe and Zn-enriched beans) but also to increase the quality of plant-based products; that is, developing plants able to accumulate minimum quantities of toxic elements such as As or Cd in edible tissues despite being grown in soils containing both, essential and non-essential metals. This can only be achieved by integrating and understanding the different processes mediating metal sensing, uptake, allocation and distribution of elements throughout the plant. Our lab uses a kinetic modeling approach (figure 1) that combines experimental data (gene expression, ionomics and radiotracer fluxes) to develop computer models describing how elements are distributed throughout the plant. These in silico models are critical to identify processes (i.e. uptake, translocation, re-mobilization figure 2) that can be modified to increase (or decrease) the delivery of certain elements to edible parts of the plant, including seeds.
references:
Mendoza-Cózatl DG, Jobe TO, Hauser F, Schroeder JI. 2011. Long-distance transport, vacuolar sequestration, tolerance, and transcriptional responses induced by cadmium and arsenic. Curr Opin Plant Biol. 14(5):554-62.
Castro-Guerrero NA, Isidra-Arellano MC, Mendoza-Cózatl DG, Valdés-López O. 2016. Common Bean: A Legume Model on the Rise for Unraveling Responses and Adaptations to Iron, Zinc, and Phosphate Deficiencies. Front Plant Sci. 7:600.